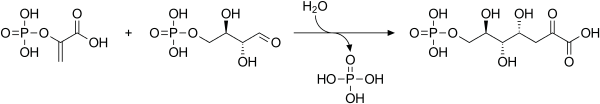3-Deoxy-D-arabino-heptulosonic acid 7-phosphate
 Fischer projection of DAHP
| |
| Names | |
|---|---|
| IUPAC name
3-Deoxy-7-O-phosphono-D-arabino-hept-2-ulosonic acid
| |
| Systematic IUPAC name
(4R,5S,6R)-4,5,6-Trihydroxy-2-oxo-7-(phosphonooxy)heptanoic acid | |
| Other names
3-Deoxy-D-arabino-heptulosonic acid 7-phosphate
3-Deoxy-D-arabino-heptulosonate-7-phosphate DAHP 7-Phospho-2-dehydro-3-deoxy-D-arabino-heptonate | |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C7H13O10P | |
| Molar mass | 288.145 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
3-Deoxy-D-arabino-heptulosonic acid 7-phosphate (DAHP) is a 7-carbon ulosonic acid. This compound is found in the shikimic acid biosynthesis pathway and is an intermediate in the production of aromatic amino acids.
Phosphoenolpyruvate and erythrose-4-phosphate react to form 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP), in a reaction catalyzed by the enzyme DAHP synthase.
DAHP is then transformed to 3-dehydroquinate (DHQ), in a reaction catalyzed by DHQ synthase. Although this reaction requires nicotinamide adenine dinucleotide (NAD) as a cofactor, the enzymic mechanism regenerates it, resulting in the net use of no NAD.

Biosynthesis of quinate catalyzed by 3-dehydroquinate synthase.
The mechanism of ring closure is complex, but involves an aldol condensation at C-2 and C-7.
Metabolic engineering has improved production of DAHP by Escherichia coli.[1] The first step, condensation of 3-deoxy-D-arabino-heptulosonic acid 7-phosphate (DAHP) from PEP/E4P, uses three isoenzymes AroF, AroG, and AroH.[2] Each one of these has its synthesis regulated from tyrosine, phenylalanine, and tryptophan, respectively. These isoenzymes all have the ability to help regulate synthesis of DAHP by the method of feedback inhibition. This acts in the cell by monitoring the concentrations of each of the three aromatic amino acids. When there is too much of any one of them, that one will allosterically control the DAHP synthetase by “turning it off”. With the first step of the common pathway shut off, synthesis of the three amino acids can not proceed. The rest of the enzymes is in the common pathway (conversion of DAHP to chorismate).
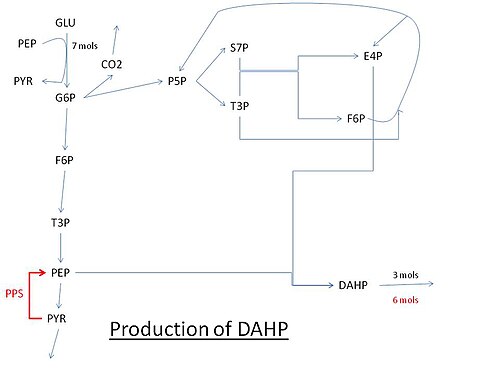
Pathway for the production of DAHP using glucose. Abbreviations: GLU: glucose, G6P: Glucose 6-phosphate, PEP: Phosphoenolpyruvate, PYR: Pyruvate, F6P: Fructose 6-phosphate, P5P: Pentose 5-phosphate (ribose or xylulose 5-phosphate) T3P: Glyceraldehyde 3-phosphate, S7P: Sedoheptulose 7-phosphate, E4P: Erythrose-4-phosphate, PPS: PEP synthase.
References[edit]
- ^ Patnaik, R. & Liao, J. (1994). "Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield". Appl. Environ. Microbiol. 60 (11): 3903–3908. Bibcode:1994ApEnM..60.3903P. doi:10.1128/AEM.60.11.3903-3908.1994. PMC 201913. PMID 7993080.
- ^ Patnaik, Ranjan; Spitzer, Richard G.; Liao, James C. (1995). "Pathway engineering for production of aromatics inEscherichia coli: Confirmation of stoichiometric analysis by independent modulation of AroG, TktA, and Pps activities". Biotechnology and Bioengineering. 46 (4): 361–70. doi:10.1002/bit.260460409. PMID 18623323. S2CID 25862511.
External links[edit]
- Webby, Celia J.; Baker, Heather M.; Lott, J. Shaun; Baker, Edward N.; Parker, Emily J. (2005). "The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis reveals a common catalytic scaffold and ancestry for type I and type II enzymes". Journal of Molecular Biology. 354 (4): 927–39. doi:10.1016/j.jmb.2005.09.093. PMID 16288916.